의연방00) 데이터 클리닉 특강
의학연구방법론 개강전 특강
📜 제목으로 보기✏마지막 댓글로
- 라이브러리 설치
- 기초 특강
- 패키지 설치 및 로드
- dply r 패키지의 파이프라인를 이용한 데이터클리닝
- 데이터클리닝
- 로드 전, 데이터를 읽기전용으로 비꿔주기
- view() 빈 데이터가 NA로 인식되는지 확인하기
- id가 유니크한지 (중복distinct.count) 확인 하기
- 목표인 summary에 넣어보기
- 변수명 긴 것부터 처리
- 날짜형으로 바꿔주기
- 파이프라인으로 클리닝과정 추가하기
- mutate() 대신 mutate_at( vars(같은작업할, 칼러들), list(~적용함수(.으로 칼럼대체)) 로 여러칼럼 한번에 처리하기
- 연속형인데 character인 애들
- replace로 문자열숫자 -> 숫자로 바꿔주기
- -결측(대문자-"NA")으로-처리해보자.">문자열"."과 "na"이가 들어가있는 숫자형 변수 -> 결측(대문자 "NA")으로 처리해보자.
- "n/a" 처리 volume_change의 문제
- 범주형인데, 연속형으로 표기되어있는 -> mutate_at</h4> local_6m , sex? </div> </div> </div> dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.) %>% mutate_at(vars(OP_date, Recur_date), list(~as.Date(., format = "%Y-%m-%d") )) %>% mutate(CA19.9=as.double(replace(dat0$CA19.9, dat0$CA19.9 == "<1.0", 1)), adc_decrease=as.double(replace( adc_decrease, adc_decrease=="na" | adc_decrease==".", NA)), # 마지막엔 더블형변환해야하니 as.double부터 volume_change=as.double(replace(volume_change, volume_change=="n/a", NA))) %>% mutate_at(vars(sex, Recur, local_6m, TNM, CCI), list(~as.factor(.))) Warning message in `[<-.factor`(`*tmp*`, list, value = 1): "invalid factor level, NA generated" summary(dat1) id age sex Recur OP_date Min. : 1.00 Min. :25.00 0:17 0:28 Min. :2011-12-29 1st Qu.:13.25 1st Qu.:49.25 1:33 1:22 1st Qu.:2015-01-28 Median :25.50 Median :58.50 Median :2015-11-08 Mean :25.50 Mean :56.44 Mean :2015-08-11 3rd Qu.:37.75 3rd Qu.:65.75 3rd Qu.:2016-09-05 Max. :50.00 Max. :79.00 Max. :2017-11-14 Recur_date local_6m CA19.9 CEA TNM CCI Min. :2012-04-09 0:27 Min. : 2.00 Min. : 0.630 1: 2 0: 3 1st Qu.:2016-06-25 1:23 1st Qu.:12.75 1st Qu.: 1.450 2: 5 1:16 Median :2018-03-13 Median :24.50 Median : 2.500 3:12 2:15 Mean :2017-09-21 Mean :24.27 Mean : 5.574 4:25 3:12 3rd Qu.:2019-10-12 3rd Qu.:35.25 3rd Qu.: 4.150 5: 6 4: 3 Max. :2020-01-11 Max. :47.00 Max. :115.900 6: 1 NA's :2 adc_decrease adc_5_decrease volume_change apt_1_mean Min. : 1.00 Min. :-6.717 Min. : 1.00 Min. : 0.628 1st Qu.:10.75 1st Qu.: 2.566 1st Qu.:12.75 1st Qu.: 1.831 Median :22.50 Median :11.329 Median :24.50 Median : 2.510 Mean :20.67 Mean :10.998 Mean :24.50 Mean : 3.129 3rd Qu.:31.00 3rd Qu.:15.638 3rd Qu.:36.25 3rd Qu.: 3.100 Max. :38.00 Max. :63.617 Max. :48.00 Max. :27.473 NA's :2 NA's :2 apt_1_95p adc_1_mean adc_1_5p Min. :1.267 : 1 Min. : 53.86 1st Qu.:2.517 100.3332748: 1 1st Qu.: 75.78 Median :3.034 100.7850723: 1 Median : 85.10 Mean :3.065 101.1288986: 1 Mean : 85.03 3rd Qu.:3.650 106.94384 : 1 3rd Qu.: 92.43 Max. :5.437 107.4796753: 1 Max. :116.59 (Other) :44 튀어는 값들(LabValue들)을 log변환 CA19.9 정상치는 거기서 거기인데, 이상인 경우 수천배로 팍 튀는 경우 -> 데이터의 분포가 너무 길어져 원본으로는 다른변수와의 관계가 깨진다. hist를 그려, 극소수만 엄청 튀는 것을 확인하자. hist(dat1$CA19.9) ㅇㅇ hist( log) log.을 붙여 새로운 변수로 추가 0이 있으면 다른 값 or 1로 바꿔서 로그취해야함. dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.) %>% mutate_at(vars(OP_date, Recur_date), list(~as.Date(., format = "%Y-%m-%d") )) %>% mutate(CA19.9=as.double(replace(dat0$CA19.9, dat0$CA19.9 == "<1.0", 1)), adc_decrease=as.double(replace( adc_decrease, adc_decrease=="na" | adc_decrease==".", NA)), volume_change=as.double(replace(volume_change, volume_change=="n/a", NA)), # 여기(mutate)다 추가 log.CA19.9 = log(CA19.9), log.CEA=log(CEA) # 위에다 추가 ) %>% mutate_at(vars(sex, Recur, local_6m, TNM, CCI), list(~as.factor(.))) Warning message in `[<-.factor`(`*tmp*`, list, value = 1): "invalid factor level, NA generated" 36분 바이닝 CA19 추가 by ifelse -> asfactor작업에도 변수 1개 추가 dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.) %>% mutate_at(vars(OP_date, Recur_date), list(~as.Date(., format = "%Y-%m-%d") )) %>% mutate(CA19.9=as.double(replace(dat0$CA19.9, dat0$CA19.9 == "<1.0", 1)), adc_decrease=as.double(replace( adc_decrease, adc_decrease=="na" | adc_decrease==".", NA)), volume_change=as.double(replace(volume_change, volume_change=="n/a", NA)), log.CA19.9 = log(CA19.9), log.CEA=log(CEA), CA19.9.group=ifelse(CA19.9>37, 1, 0), # 칼럼 생성 ) %>% mutate_at(vars(sex, Recur, local_6m, TNM, CCI, CA19.9 # factor화 추가 ), list(~as.factor(.))) Warning message in `[<-.factor`(`*tmp*`, list, value = 1): "invalid factor level, NA generated" ply r 을 통해 mapvalue (1,2를 묶어 -> TNM.b칼럼으로)쓰기 from 원래 카테고리 -> to = 합칠거명 같은이름으로 순서대로 차례대로 카테고리 ) 다시 범주형 만드는 곳에서 처리 dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.) %>% mutate_at(vars(OP_date, Recur_date), list(~as.Date(., format = "%Y-%m-%d") )) %>% mutate(CA19.9=as.double(replace(dat0$CA19.9, dat0$CA19.9 == "<1.0", 1)), adc_decrease=as.double(replace( adc_decrease, adc_decrease=="na" | adc_decrease==".", NA)), volume_change=as.double(replace(volume_change, volume_change=="n/a", NA)), log.CA19.9 = log(CA19.9), log.CEA=log(CEA), CA19.9.group=ifelse(CA19.9>37, 1, 0), TNM.b = mapvalues(TNM, from=c(1,2,3,4,5), to=c("1or2", "1or2", "3", "4", "5")), # 카테고리 합치기 ) %>% mutate_at(vars(sex, Recur, local_6m, TNM, CCI, CA19.9, TNM.b # 추가 ), list(~as.factor(.))) Warning message in `[<-.factor`(`*tmp*`, list, value = 1): "invalid factor level, NA generated" dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.) %>% mutate_at(vars(OP_date, Recur_date), list(~as.Date(., format = "%Y-%m-%d") )) %>% mutate(CA19.9=as.double(replace(dat0$CA19.9, dat0$CA19.9 == "<1.0", 1)), adc_decrease=as.double(replace( adc_decrease, adc_decrease=="na" | adc_decrease==".", NA)), volume_change=as.double(replace(volume_change, volume_change=="n/a", NA)), log.CA19.9 = log(CA19.9), log.CEA=log(CEA), CA19.9.group=ifelse(CA19.9>37, 1, 0), TNM.b = mapvalues(TNM, from=c(1,2,3,4,5), to=c("1or2", "1or2", "3", "4", "5")), CCI.b = mapvalues(CCI, from=c(0,1,2,3,4,6), to=c("0or1", "0or1", "2", "3", ">=4", ">=4")), ) %>% mutate_at(vars(sex, Recur, local_6m, TNM, CCI, CA19.9, TNM.b # 추가 ), list(~as.factor(.))) Warning message in `[<-.factor`(`*tmp*`, list, value = 1): "invalid factor level, NA generated" 여기서부터는 못따라감 # as.factor가 아닌 factor로 어떤 순서대로 카테고리를 하고싶은지 인식시켜준다 # 아래에서 as.factor부분에서 빼준다. 45분부터 못따라감.. 48까지 클리닝은 끝 library("tableone") tableone으로 쉽게 table만들기 var1<-c("Recur", "RFS", "local_6m", "age", "sex", "CA19.9", "log.CA19.9", "CA19.9.group", "CEA", "log.CEA", "TNM", "TNM.b", "CCI", "CCI.b", "adc_decrease", "adc_5_decrease", "volume_change", "apt_1_mean", "apt_1_95p", "adc_1_mean", "adc_1_5p") tobj<-CreateTableOne(vars = var1, data=dat1) summary(tobj) Warning message in ModuleReturnVarsExist(vars, data): "The data frame does not have: RFS Dropped" ### Summary of continuous variables ### strata: Overall n miss p.miss mean sd median p25 p75 min max skew kurt age 50 0 0 56.4 12.6 58.5 49.2 66 25.0 79 -0.47 -0.3 log.CA19.9 50 2 4 2.9 0.8 3.2 2.5 4 0.7 4 -1.12 0.5 CA19.9.group 50 2 4 0.2 0.4 0.0 0.0 0 0.0 1 1.48 0.2 CEA 50 0 0 5.6 16.3 2.5 1.4 4 0.6 116 6.63 45.5 log.CEA 50 0 0 1.0 0.9 0.9 0.4 1 -0.5 5 1.81 5.8 adc_decrease 50 2 4 20.7 11.5 22.5 10.8 31 1.0 38 -0.31 -1.3 adc_5_decrease 50 0 0 11.0 12.2 11.3 2.6 16 -6.7 64 1.74 6.0 volume_change 50 2 4 24.5 14.0 24.5 12.8 36 1.0 48 0.00 -1.2 apt_1_mean 50 0 0 3.1 3.7 2.5 1.8 3 0.6 27 6.06 40.2 apt_1_95p 50 0 0 3.1 0.9 3.0 2.5 4 1.3 5 0.40 0.5 adc_1_5p 50 0 0 85.0 11.5 85.1 75.8 92 53.9 117 0.05 0.5 ======================================================================================= ### Summary of categorical variables ### strata: Overall var n miss p.miss level freq percent cum.percent Recur 50 0 0.0 0 28 56.0 56.0 1 22 44.0 100.0 local_6m 50 0 0.0 0 27 54.0 54.0 1 23 46.0 100.0 sex 50 0 0.0 0 17 34.0 34.0 1 33 66.0 100.0 CA19.9 50 2 4.0 2 1 2.1 2.1 3 2 4.2 6.2 4 1 2.1 8.3 5 1 2.1 10.4 6 1 2.1 12.5 7 1 2.1 14.6 8 1 2.1 16.7 9 1 2.1 18.8 10 1 2.1 20.8 11 1 2.1 22.9 12 1 2.1 25.0 13 1 2.1 27.1 14 1 2.1 29.2 15 1 2.1 31.2 16 1 2.1 33.3 17 1 2.1 35.4 18 1 2.1 37.5 19 1 2.1 39.6 20 1 2.1 41.7 21 1 2.1 43.8 22 1 2.1 45.8 23 1 2.1 47.9 24 1 2.1 50.0 25 1 2.1 52.1 26 1 2.1 54.2 27 1 2.1 56.2 28 1 2.1 58.3 29 1 2.1 60.4 30 1 2.1 62.5 31 1 2.1 64.6 32 1 2.1 66.7 33 1 2.1 68.8 34 1 2.1 70.8 35 2 4.2 75.0 36 1 2.1 77.1 37 1 2.1 79.2 38 1 2.1 81.2 39 1 2.1 83.3 40 1 2.1 85.4 41 1 2.1 87.5 42 1 2.1 89.6 43 1 2.1 91.7 44 1 2.1 93.8 45 1 2.1 95.8 46 1 2.1 97.9 47 1 2.1 100.0 TNM 50 0 0.0 1 2 4.0 4.0 2 5 10.0 14.0 3 12 24.0 38.0 4 25 50.0 88.0 5 6 12.0 100.0 TNM.b 50 0 0.0 1or2 7 14.0 14.0 3 12 24.0 38.0 4 25 50.0 88.0 5 6 12.0 100.0 CCI 50 0 0.0 0 3 6.0 6.0 1 16 32.0 38.0 2 15 30.0 68.0 3 12 24.0 92.0 4 3 6.0 98.0 6 1 2.0 100.0 CCI.b 50 0 0.0 >=4 4 8.0 8.0 0or1 19 38.0 46.0 2 15 30.0 76.0 3 12 24.0 100.0 adc_1_mean 50 0 0.0 1 2.0 2.0 100.3332748 1 2.0 4.0 100.7850723 1 2.0 6.0 101.1288986 1 2.0 8.0 106.94384 1 2.0 10.0 107.4796753 1 2.0 12.0 108.1219025 1 2.0 14.0 108.4807739 1 2.0 16.0 109.8269577 1 2.0 18.0 110.6593323 1 2.0 20.0 112.250946 1 2.0 22.0 112.6311264 1 2.0 24.0 113.6343842 1 2.0 26.0 114.5146027 1 2.0 28.0 116.6255951 1 2.0 30.0 116.9603348 1 2.0 32.0 119.7995605 1 2.0 34.0 121.1716843 1 2.0 36.0 123.4509964 1 2.0 38.0 123.5778046 1 2.0 40.0 125.6047516 1 2.0 42.0 125.8717575 1 2.0 44.0 127.4504852 1 2.0 46.0 128.1634064 1 2.0 48.0 128.2574005 1 2.0 50.0 128.8485718 1 2.0 52.0 129.5490875 1 2.0 54.0 129.7754059 1 2.0 56.0 130.4454651 1 2.0 58.0 131.0157623 1 2.0 60.0 131.2865448 1 2.0 62.0 133.0798645 1 2.0 64.0 134.7369385 1 2.0 66.0 137.2429199 1 2.0 68.0 137.4812317 1 2.0 70.0 138.7712097 1 2.0 72.0 139.8694763 1 2.0 74.0 140.6768646 1 2.0 76.0 142.7576294 1 2.0 78.0 144.4676361 1 2.0 80.0 144.8905334 1 2.0 82.0 146.3157654 1 2.0 84.0 149.0292358 1 2.0 86.0 152.9427185 1 2.0 88.0 154.0635529 1 2.0 90.0 154.9950104 1 2.0 92.0 82.6672821 1 2.0 94.0 89.37554932 1 2.0 96.0 99.98454285 1 2.0 98.0 n/a 1 2.0 100.0 # 범주형 변수들의 요약통계량이 나온다. 갯수와 % print(tobj) Overall n 50 Recur = 1 (%) 22 (44.0) local_6m = 1 (%) 23 (46.0) age (mean (SD)) 56.44 (12.60) sex = 1 (%) 33 (66.0) CA19.9 (%) 2 1 ( 2.1) 3 2 ( 4.2) 4 1 ( 2.1) 5 1 ( 2.1) 6 1 ( 2.1) 7 1 ( 2.1) 8 1 ( 2.1) 9 1 ( 2.1) 10 1 ( 2.1) 11 1 ( 2.1) 12 1 ( 2.1) 13 1 ( 2.1) 14 1 ( 2.1) 15 1 ( 2.1) 16 1 ( 2.1) 17 1 ( 2.1) 18 1 ( 2.1) 19 1 ( 2.1) 20 1 ( 2.1) 21 1 ( 2.1) 22 1 ( 2.1) 23 1 ( 2.1) 24 1 ( 2.1) 25 1 ( 2.1) 26 1 ( 2.1) 27 1 ( 2.1) 28 1 ( 2.1) 29 1 ( 2.1) 30 1 ( 2.1) 31 1 ( 2.1) 32 1 ( 2.1) 33 1 ( 2.1) 34 1 ( 2.1) 35 2 ( 4.2) 36 1 ( 2.1) 37 1 ( 2.1) 38 1 ( 2.1) 39 1 ( 2.1) 40 1 ( 2.1) 41 1 ( 2.1) 42 1 ( 2.1) 43 1 ( 2.1) 44 1 ( 2.1) 45 1 ( 2.1) 46 1 ( 2.1) 47 1 ( 2.1) log.CA19.9 (mean (SD)) 2.95 (0.81) CA19.9.group (mean (SD)) 0.21 (0.41) CEA (mean (SD)) 5.57 (16.28) log.CEA (mean (SD)) 0.99 (0.89) TNM (%) 1 2 ( 4.0) 2 5 (10.0) 3 12 (24.0) 4 25 (50.0) 5 6 (12.0) TNM.b (%) 1or2 7 (14.0) 3 12 (24.0) 4 25 (50.0) 5 6 (12.0) CCI (%) 0 3 ( 6.0) 1 16 (32.0) 2 15 (30.0) 3 12 (24.0) 4 3 ( 6.0) 6 1 ( 2.0) CCI.b (%) >=4 4 ( 8.0) 0or1 19 (38.0) 2 15 (30.0) 3 12 (24.0) adc_decrease (mean (SD)) 20.67 (11.49) adc_5_decrease (mean (SD)) 11.00 (12.19) volume_change (mean (SD)) 24.50 (14.00) apt_1_mean (mean (SD)) 3.13 (3.70) apt_1_95p (mean (SD)) 3.07 (0.88) adc_1_mean (%) 1 ( 2.0) 100.3332748 1 ( 2.0) 100.7850723 1 ( 2.0) 101.1288986 1 ( 2.0) 106.94384 1 ( 2.0) 107.4796753 1 ( 2.0) 108.1219025 1 ( 2.0) 108.4807739 1 ( 2.0) 109.8269577 1 ( 2.0) 110.6593323 1 ( 2.0) 112.250946 1 ( 2.0) 112.6311264 1 ( 2.0) 113.6343842 1 ( 2.0) 114.5146027 1 ( 2.0) 116.6255951 1 ( 2.0) 116.9603348 1 ( 2.0) 119.7995605 1 ( 2.0) 121.1716843 1 ( 2.0) 123.4509964 1 ( 2.0) 123.5778046 1 ( 2.0) 125.6047516 1 ( 2.0) 125.8717575 1 ( 2.0) 127.4504852 1 ( 2.0) 128.1634064 1 ( 2.0) 128.2574005 1 ( 2.0) 128.8485718 1 ( 2.0) 129.5490875 1 ( 2.0) 129.7754059 1 ( 2.0) 130.4454651 1 ( 2.0) 131.0157623 1 ( 2.0) 131.2865448 1 ( 2.0) 133.0798645 1 ( 2.0) 134.7369385 1 ( 2.0) 137.2429199 1 ( 2.0) 137.4812317 1 ( 2.0) 138.7712097 1 ( 2.0) 139.8694763 1 ( 2.0) 140.6768646 1 ( 2.0) 142.7576294 1 ( 2.0) 144.4676361 1 ( 2.0) 144.8905334 1 ( 2.0) 146.3157654 1 ( 2.0) 149.0292358 1 ( 2.0) 152.9427185 1 ( 2.0) 154.0635529 1 ( 2.0) 154.9950104 1 ( 2.0) 82.6672821 1 ( 2.0) 89.37554932 1 ( 2.0) 99.98454285 1 ( 2.0) n/a 1 ( 2.0) adc_1_5p (mean (SD)) 85.03 (11.48) print(tobj, showAllLevels = T) level Overall n 50 Recur (%) 0 28 (56.0) 1 22 (44.0) local_6m (%) 0 27 (54.0) 1 23 (46.0) age (mean (SD)) 56.44 (12.60) sex (%) 0 17 (34.0) 1 33 (66.0) CA19.9 (%) 2 1 ( 2.1) 3 2 ( 4.2) 4 1 ( 2.1) 5 1 ( 2.1) 6 1 ( 2.1) 7 1 ( 2.1) 8 1 ( 2.1) 9 1 ( 2.1) 10 1 ( 2.1) 11 1 ( 2.1) 12 1 ( 2.1) 13 1 ( 2.1) 14 1 ( 2.1) 15 1 ( 2.1) 16 1 ( 2.1) 17 1 ( 2.1) 18 1 ( 2.1) 19 1 ( 2.1) 20 1 ( 2.1) 21 1 ( 2.1) 22 1 ( 2.1) 23 1 ( 2.1) 24 1 ( 2.1) 25 1 ( 2.1) 26 1 ( 2.1) 27 1 ( 2.1) 28 1 ( 2.1) 29 1 ( 2.1) 30 1 ( 2.1) 31 1 ( 2.1) 32 1 ( 2.1) 33 1 ( 2.1) 34 1 ( 2.1) 35 2 ( 4.2) 36 1 ( 2.1) 37 1 ( 2.1) 38 1 ( 2.1) 39 1 ( 2.1) 40 1 ( 2.1) 41 1 ( 2.1) 42 1 ( 2.1) 43 1 ( 2.1) 44 1 ( 2.1) 45 1 ( 2.1) 46 1 ( 2.1) 47 1 ( 2.1) log.CA19.9 (mean (SD)) 2.95 (0.81) CA19.9.group (mean (SD)) 0.21 (0.41) CEA (mean (SD)) 5.57 (16.28) log.CEA (mean (SD)) 0.99 (0.89) TNM (%) 1 2 ( 4.0) 2 5 (10.0) 3 12 (24.0) 4 25 (50.0) 5 6 (12.0) TNM.b (%) 1or2 7 (14.0) 3 12 (24.0) 4 25 (50.0) 5 6 (12.0) CCI (%) 0 3 ( 6.0) 1 16 (32.0) 2 15 (30.0) 3 12 (24.0) 4 3 ( 6.0) 6 1 ( 2.0) CCI.b (%) >=4 4 ( 8.0) 0or1 19 (38.0) 2 15 (30.0) 3 12 (24.0) adc_decrease (mean (SD)) 20.67 (11.49) adc_5_decrease (mean (SD)) 11.00 (12.19) volume_change (mean (SD)) 24.50 (14.00) apt_1_mean (mean (SD)) 3.13 (3.70) apt_1_95p (mean (SD)) 3.07 (0.88) adc_1_mean (%) 1 ( 2.0) 100.3332748 1 ( 2.0) 100.7850723 1 ( 2.0) 101.1288986 1 ( 2.0) 106.94384 1 ( 2.0) 107.4796753 1 ( 2.0) 108.1219025 1 ( 2.0) 108.4807739 1 ( 2.0) 109.8269577 1 ( 2.0) 110.6593323 1 ( 2.0) 112.250946 1 ( 2.0) 112.6311264 1 ( 2.0) 113.6343842 1 ( 2.0) 114.5146027 1 ( 2.0) 116.6255951 1 ( 2.0) 116.9603348 1 ( 2.0) 119.7995605 1 ( 2.0) 121.1716843 1 ( 2.0) 123.4509964 1 ( 2.0) 123.5778046 1 ( 2.0) 125.6047516 1 ( 2.0) 125.8717575 1 ( 2.0) 127.4504852 1 ( 2.0) 128.1634064 1 ( 2.0) 128.2574005 1 ( 2.0) 128.8485718 1 ( 2.0) 129.5490875 1 ( 2.0) 129.7754059 1 ( 2.0) 130.4454651 1 ( 2.0) 131.0157623 1 ( 2.0) 131.2865448 1 ( 2.0) 133.0798645 1 ( 2.0) 134.7369385 1 ( 2.0) 137.2429199 1 ( 2.0) 137.4812317 1 ( 2.0) 138.7712097 1 ( 2.0) 139.8694763 1 ( 2.0) 140.6768646 1 ( 2.0) 142.7576294 1 ( 2.0) 144.4676361 1 ( 2.0) 144.8905334 1 ( 2.0) 146.3157654 1 ( 2.0) 149.0292358 1 ( 2.0) 152.9427185 1 ( 2.0) 154.0635529 1 ( 2.0) 154.9950104 1 ( 2.0) 82.6672821 1 ( 2.0) 89.37554932 1 ( 2.0) 99.98454285 1 ( 2.0) n/a 1 ( 2.0) adc_1_5p (mean (SD)) 85.03 (11.48) # 하지만 치우친 [기울어진 분포]의 연속형변수는 [ median, IQR ]를 뽑아야한다. print(tobj, showAllLevels = T, nonnormal = c("CA19.9", "CEA")) level Overall n 50 Recur (%) 0 28 (56.0) 1 22 (44.0) local_6m (%) 0 27 (54.0) 1 23 (46.0) age (mean (SD)) 56.44 (12.60) sex (%) 0 17 (34.0) 1 33 (66.0) CA19.9 (%) 2 1 ( 2.1) 3 2 ( 4.2) 4 1 ( 2.1) 5 1 ( 2.1) 6 1 ( 2.1) 7 1 ( 2.1) 8 1 ( 2.1) 9 1 ( 2.1) 10 1 ( 2.1) 11 1 ( 2.1) 12 1 ( 2.1) 13 1 ( 2.1) 14 1 ( 2.1) 15 1 ( 2.1) 16 1 ( 2.1) 17 1 ( 2.1) 18 1 ( 2.1) 19 1 ( 2.1) 20 1 ( 2.1) 21 1 ( 2.1) 22 1 ( 2.1) 23 1 ( 2.1) 24 1 ( 2.1) 25 1 ( 2.1) 26 1 ( 2.1) 27 1 ( 2.1) 28 1 ( 2.1) 29 1 ( 2.1) 30 1 ( 2.1) 31 1 ( 2.1) 32 1 ( 2.1) 33 1 ( 2.1) 34 1 ( 2.1) 35 2 ( 4.2) 36 1 ( 2.1) 37 1 ( 2.1) 38 1 ( 2.1) 39 1 ( 2.1) 40 1 ( 2.1) 41 1 ( 2.1) 42 1 ( 2.1) 43 1 ( 2.1) 44 1 ( 2.1) 45 1 ( 2.1) 46 1 ( 2.1) 47 1 ( 2.1) log.CA19.9 (mean (SD)) 2.95 (0.81) CA19.9.group (mean (SD)) 0.21 (0.41) CEA (median [IQR]) 2.50 [1.45, 4.15] log.CEA (mean (SD)) 0.99 (0.89) TNM (%) 1 2 ( 4.0) 2 5 (10.0) 3 12 (24.0) 4 25 (50.0) 5 6 (12.0) TNM.b (%) 1or2 7 (14.0) 3 12 (24.0) 4 25 (50.0) 5 6 (12.0) CCI (%) 0 3 ( 6.0) 1 16 (32.0) 2 15 (30.0) 3 12 (24.0) 4 3 ( 6.0) 6 1 ( 2.0) CCI.b (%) >=4 4 ( 8.0) 0or1 19 (38.0) 2 15 (30.0) 3 12 (24.0) adc_decrease (mean (SD)) 20.67 (11.49) adc_5_decrease (mean (SD)) 11.00 (12.19) volume_change (mean (SD)) 24.50 (14.00) apt_1_mean (mean (SD)) 3.13 (3.70) apt_1_95p (mean (SD)) 3.07 (0.88) adc_1_mean (%) 1 ( 2.0) 100.3332748 1 ( 2.0) 100.7850723 1 ( 2.0) 101.1288986 1 ( 2.0) 106.94384 1 ( 2.0) 107.4796753 1 ( 2.0) 108.1219025 1 ( 2.0) 108.4807739 1 ( 2.0) 109.8269577 1 ( 2.0) 110.6593323 1 ( 2.0) 112.250946 1 ( 2.0) 112.6311264 1 ( 2.0) 113.6343842 1 ( 2.0) 114.5146027 1 ( 2.0) 116.6255951 1 ( 2.0) 116.9603348 1 ( 2.0) 119.7995605 1 ( 2.0) 121.1716843 1 ( 2.0) 123.4509964 1 ( 2.0) 123.5778046 1 ( 2.0) 125.6047516 1 ( 2.0) 125.8717575 1 ( 2.0) 127.4504852 1 ( 2.0) 128.1634064 1 ( 2.0) 128.2574005 1 ( 2.0) 128.8485718 1 ( 2.0) 129.5490875 1 ( 2.0) 129.7754059 1 ( 2.0) 130.4454651 1 ( 2.0) 131.0157623 1 ( 2.0) 131.2865448 1 ( 2.0) 133.0798645 1 ( 2.0) 134.7369385 1 ( 2.0) 137.2429199 1 ( 2.0) 137.4812317 1 ( 2.0) 138.7712097 1 ( 2.0) 139.8694763 1 ( 2.0) 140.6768646 1 ( 2.0) 142.7576294 1 ( 2.0) 144.4676361 1 ( 2.0) 144.8905334 1 ( 2.0) 146.3157654 1 ( 2.0) 149.0292358 1 ( 2.0) 152.9427185 1 ( 2.0) 154.0635529 1 ( 2.0) 154.9950104 1 ( 2.0) 82.6672821 1 ( 2.0) 89.37554932 1 ( 2.0) 99.98454285 1 ( 2.0) n/a 1 ( 2.0) adc_1_5p (mean (SD)) 85.03 (11.48) ?CreateTableOne # createtableone에서 strata에 그룹을 나눈 칼럼명 options(width = 300) tobj2<-CreateTableOne(vars = var1, data=dat1, strata="CA19.9.group") print(tobj2) Warning message in ModuleReturnVarsExist(vars, data): "The data frame does not have: RFS Dropped" Stratified by CA19.9.group 0 1 p test n 38 10 Recur = 1 (%) 15 (39.5) 7 (70.0) 0.172 local_6m = 1 (%) 20 (52.6) 3 (30.0) 0.358 age (mean (SD)) 56.34 (12.56) 58.80 (13.62) 0.591 sex = 1 (%) 24 (63.2) 9 (90.0) 0.213 CA19.9 (%) 0.352 2 1 ( 2.6) 0 ( 0.0) 3 2 ( 5.3) 0 ( 0.0) 4 1 ( 2.6) 0 ( 0.0) 5 1 ( 2.6) 0 ( 0.0) 6 1 ( 2.6) 0 ( 0.0) 7 1 ( 2.6) 0 ( 0.0) 8 1 ( 2.6) 0 ( 0.0) 9 1 ( 2.6) 0 ( 0.0) 10 1 ( 2.6) 0 ( 0.0) 11 1 ( 2.6) 0 ( 0.0) 12 1 ( 2.6) 0 ( 0.0) 13 1 ( 2.6) 0 ( 0.0) 14 1 ( 2.6) 0 ( 0.0) 15 1 ( 2.6) 0 ( 0.0) 16 1 ( 2.6) 0 ( 0.0) 17 1 ( 2.6) 0 ( 0.0) 18 1 ( 2.6) 0 ( 0.0) 19 1 ( 2.6) 0 ( 0.0) 20 1 ( 2.6) 0 ( 0.0) 21 1 ( 2.6) 0 ( 0.0) 22 1 ( 2.6) 0 ( 0.0) 23 1 ( 2.6) 0 ( 0.0) 24 1 ( 2.6) 0 ( 0.0) 25 1 ( 2.6) 0 ( 0.0) 26 1 ( 2.6) 0 ( 0.0) 27 1 ( 2.6) 0 ( 0.0) 28 1 ( 2.6) 0 ( 0.0) 29 1 ( 2.6) 0 ( 0.0) 30 1 ( 2.6) 0 ( 0.0) 31 1 ( 2.6) 0 ( 0.0) 32 1 ( 2.6) 0 ( 0.0) 33 1 ( 2.6) 0 ( 0.0) 34 1 ( 2.6) 0 ( 0.0) 35 2 ( 5.3) 0 ( 0.0) 36 1 ( 2.6) 0 ( 0.0) 37 1 ( 2.6) 0 ( 0.0) 38 0 ( 0.0) 1 (10.0) 39 0 ( 0.0) 1 (10.0) 40 0 ( 0.0) 1 (10.0) 41 0 ( 0.0) 1 (10.0) 42 0 ( 0.0) 1 (10.0) 43 0 ( 0.0) 1 (10.0) 44 0 ( 0.0) 1 (10.0) 45 0 ( 0.0) 1 (10.0) 46 0 ( 0.0) 1 (10.0) 47 0 ( 0.0) 1 (10.0) log.CA19.9 (mean (SD)) 2.74 (0.79) 3.75 (0.07) <0.001 CA19.9.group (mean (SD)) 0.00 (0.00) 1.00 (0.00) <0.001 CEA (mean (SD)) 6.13 (18.55) 4.15 (4.59) 0.740 log.CEA (mean (SD)) 0.99 (0.90) 1.03 (0.93) 0.926 TNM (%) 0.308 1 1 ( 2.6) 1 (10.0) 2 3 ( 7.9) 1 (10.0) 3 11 (28.9) 1 (10.0) 4 17 (44.7) 7 (70.0) 5 6 (15.8) 0 ( 0.0) TNM.b (%) 0.231 1or2 4 (10.5) 2 (20.0) 3 11 (28.9) 1 (10.0) 4 17 (44.7) 7 (70.0) 5 6 (15.8) 0 ( 0.0) CCI (%) 0.304 0 1 ( 2.6) 2 (20.0) 1 13 (34.2) 2 (20.0) 2 10 (26.3) 4 (40.0) 3 10 (26.3) 2 (20.0) 4 3 ( 7.9) 0 ( 0.0) 6 1 ( 2.6) 0 ( 0.0) CCI.b (%) 0.635 >=4 4 (10.5) 0 ( 0.0) 0or1 14 (36.8) 4 (40.0) 2 10 (26.3) 4 (40.0) 3 10 (26.3) 2 (20.0) adc_decrease (mean (SD)) 20.83 (11.55) 21.60 (11.41) 0.853 adc_5_decrease (mean (SD)) 11.24 (11.82) 10.80 (14.62) 0.922 volume_change (mean (SD)) 22.58 (13.56) 32.00 (13.00) 0.056 apt_1_mean (mean (SD)) 3.19 (4.20) 3.08 (1.23) 0.935 apt_1_95p (mean (SD)) 2.96 (0.75) 3.54 (1.25) 0.064 adc_1_mean (%) NaN 1 ( 2.6) 0 ( 0.0) 100.3332748 1 ( 2.6) 0 ( 0.0) 100.7850723 1 ( 2.6) 0 ( 0.0) 101.1288986 1 ( 2.6) 0 ( 0.0) 106.94384 0 ( 0.0) 1 (10.0) 107.4796753 1 ( 2.6) 0 ( 0.0) 108.1219025 0 ( 0.0) 1 (10.0) 108.4807739 1 ( 2.6) 0 ( 0.0) 109.8269577 0 ( 0.0) 0 ( 0.0) 110.6593323 1 ( 2.6) 0 ( 0.0) 112.250946 0 ( 0.0) 1 (10.0) 112.6311264 1 ( 2.6) 0 ( 0.0) 113.6343842 1 ( 2.6) 0 ( 0.0) 114.5146027 0 ( 0.0) 1 (10.0) 116.6255951 1 ( 2.6) 0 ( 0.0) 116.9603348 1 ( 2.6) 0 ( 0.0) 119.7995605 1 ( 2.6) 0 ( 0.0) 121.1716843 1 ( 2.6) 0 ( 0.0) 123.4509964 1 ( 2.6) 0 ( 0.0) 123.5778046 1 ( 2.6) 0 ( 0.0) 125.6047516 1 ( 2.6) 0 ( 0.0) 125.8717575 1 ( 2.6) 0 ( 0.0) 127.4504852 1 ( 2.6) 0 ( 0.0) 128.1634064 1 ( 2.6) 0 ( 0.0) 128.2574005 0 ( 0.0) 1 (10.0) 128.8485718 0 ( 0.0) 0 ( 0.0) 129.5490875 1 ( 2.6) 0 ( 0.0) 129.7754059 1 ( 2.6) 0 ( 0.0) 130.4454651 1 ( 2.6) 0 ( 0.0) 131.0157623 1 ( 2.6) 0 ( 0.0) 131.2865448 1 ( 2.6) 0 ( 0.0) 133.0798645 1 ( 2.6) 0 ( 0.0) 134.7369385 1 ( 2.6) 0 ( 0.0) 137.2429199 1 ( 2.6) 0 ( 0.0) 137.4812317 1 ( 2.6) 0 ( 0.0) 138.7712097 1 ( 2.6) 0 ( 0.0) 139.8694763 0 ( 0.0) 1 (10.0) 140.6768646 1 ( 2.6) 0 ( 0.0) 142.7576294 1 ( 2.6) 0 ( 0.0) 144.4676361 0 ( 0.0) 1 (10.0) 144.8905334 1 ( 2.6) 0 ( 0.0) 146.3157654 0 ( 0.0) 1 (10.0) 149.0292358 0 ( 0.0) 1 (10.0) 152.9427185 1 ( 2.6) 0 ( 0.0) 154.0635529 1 ( 2.6) 0 ( 0.0) 154.9950104 0 ( 0.0) 1 (10.0) 82.6672821 1 ( 2.6) 0 ( 0.0) 89.37554932 1 ( 2.6) 0 ( 0.0) 99.98454285 1 ( 2.6) 0 ( 0.0) n/a 1 ( 2.6) 0 ( 0.0) adc_1_5p (mean (SD)) 84.85 (11.42) 84.70 (12.29) 0.972 </div> 같은작업들>
- 튀어는 값들(LabValue들)을 log변환
- ㅇㅇ
- 36분 바이닝 CA19 추가 by ifelse -> asfactor작업에도 변수 1개 추가
- ply r 을 통해 mapvalue (1,2를 묶어 -> TNM.b칼럼으로)쓰기
- 48까지 클리닝은 끝
- tableone으로 쉽게 table만들기
- 강의 교재 사이트: https://rpubs.com/biostat81/DataPrep_CRM
- 데이터셋 보유되어있음(data/특강/example_dataset.csv)
install.packages('dplyr')
install.packages('tableone')
library(plyr)
library(dplyr)
library(tableone)
aa <- 1:5 # 벡터
bb <- as.factor(aa) # 벡터 -> factor
cc <- as.character(aa) # 벡터 -> 케릭터
bb
# factor형 변수
# levels라는 한줄이 더 나온다.
# 범주형변수 factor로 인식시켜줘야, 범주의 갯수와 종류를 print해준다.
cc
# 문자형이라서 큰따옴표로 나온다 in R studio
aa # numeric은 가감승제가 가능하다.
aa + 1 # 더하면 벡터 전체에 일괄 적용 된다.
bb + 1 # 팩터형 변수에는 가감승제를 해줄 수 없다.
cc + 1 # 케릭터형 변수에는 가감승제를 할 수 없다.
typeof(aa)
typeof(bb) # factor형은 내부에서 integer로 저장한다.
typeof(cc)
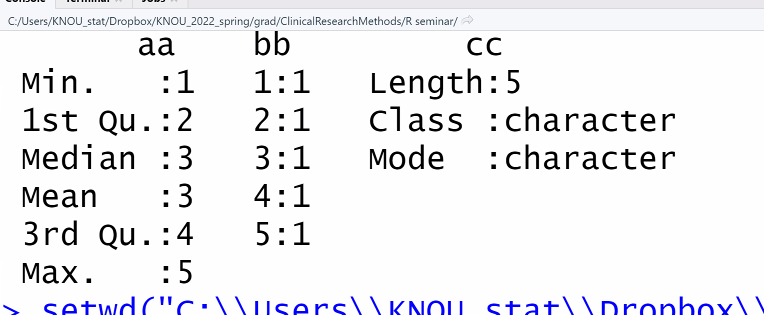
dd <- data.frame( aa, bb, cc) # 개별 벡터들을 넣어주면, 변수=>칼럼이 된다.
dd
# data.frame은 type이 달라도 모아서 사용할 수 있다.
summary(dd)
# aa: 연속형변수 -> 기본? 통계가 잘 나온다
# bb: 각 범주에 갯수를 표시한 범주형 변수 -> 도수분표표
# cc: 캐릭터변수 -> 길이에다가 설명
# 데이터클리닝 최종목표 : summary에 넣었을 때 정상적으로 나와야함.
aa
log(aa)
aa %>% log()
# 체이닝해서.. 여러가지 작업을 할 경우, 그 과정을 알 수 있다.
round( sum( log(aa) ) )
# 왼쪽부터 읽는 것 <-> 호출 순서가 다른 것을
# 파이프라인으로 순서대로 읽고 실행되게
aa %>% log() %>% sum() %>% round()
# 데이터를 객체에 저장
dat0 <- read.csv('./data/특강//example_dataset.csv')
# stringAsFactors = F) # R버전 4이하인 경우 수행해야함.
head(dat0)
view(dat0)
# 엑셀 저장 실수로 빈 데이터가 NA로 차있다. -> 50행까지만 읽자.
# dat0 <- read.csv('./data/특강//example_dataset.csv')
# 빈 데이터는 제끼기
dat0 <- read.csv('./data/특강//example_dataset.csv', nrow=50)
head(dat0)
length(unique(dat0$id))
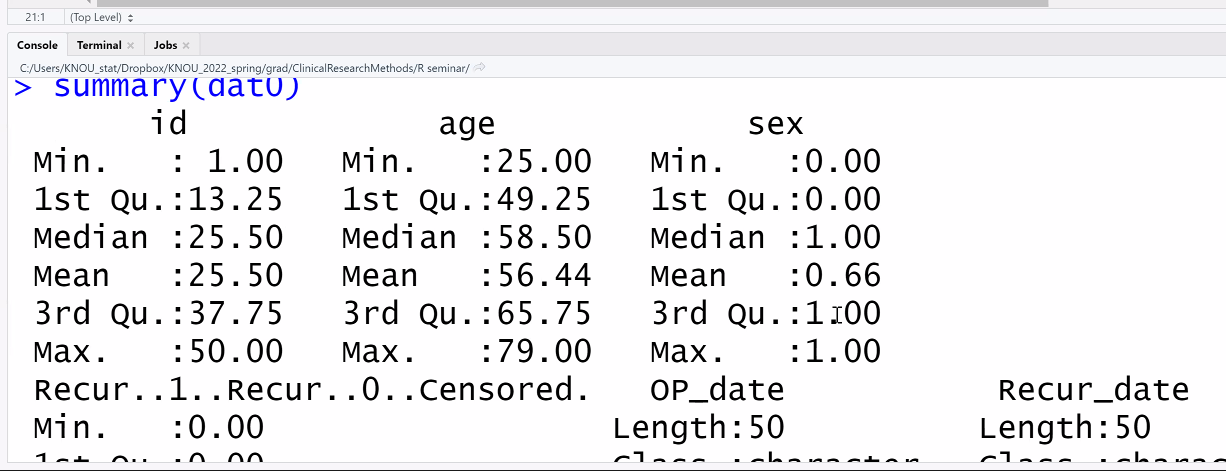
summary(dat0)
# Recur -> 변수명 짧게
# 날짜 -> character로 인식함
#local_6m
# ca19.9 -> 케릭터로 인식됨.
dat0$Recur..1..Recur..0..Censored.
# 클리닝하명 dat1에 <-
dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.)
dat0$OP_date
# 이것으로 확인한다. -> 안되면 케릭터로 잘못 된 것임
dat0$OP_date + 1
as.Date(dat0$OP_date, format = "%Y-%m-%d") + 1
# 에러 안나면 성공임.
# mutate( = )덮어써주기
dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.) %>%
mutate(OP_date=as.Date(OP_date, format = "%Y-%m-%d")) # + 1)
dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.) %>%
mutate_at(vars(OP_date, Recur_date),
list(~as.Date(., format = "%Y-%m-%d") ))
summary(dat1)
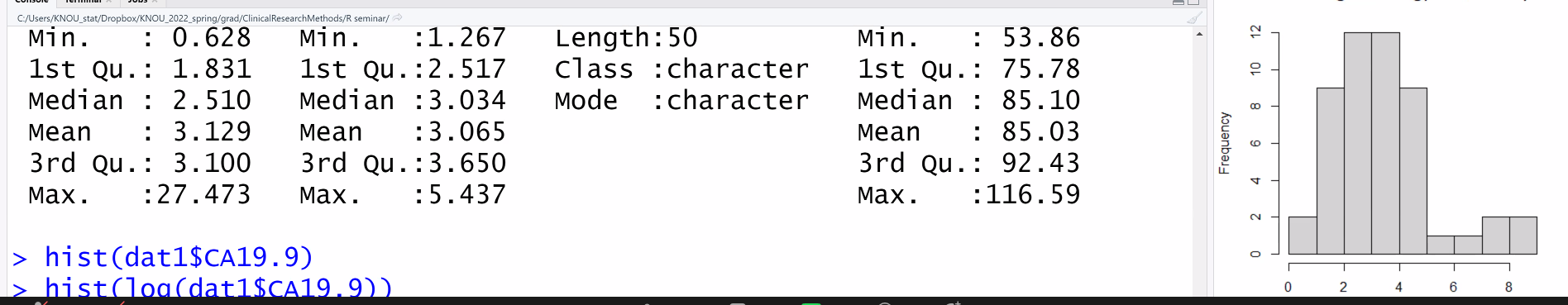
dat0$CA19.9 # 피검사 랩value
# ~보다 작다(<1)로 입력되는 경우가 많은데, 숫자가 아닌
replace(dat0$CA19.9, dat0$CA19.9 == "<1.0", 1)#위치 #바꿀값
# 숫자로 바꿔졌찌만, 칼럼 전체가 케릭터(팩터)라 "1"로 들어갈 듯
# 칼럼 전체가 응답된다.
as.double(replace(dat0$CA19.9, dat0$CA19.9 == "<1.0", 1))
dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.) %>%
mutate_at(vars(OP_date, Recur_date),
list(~as.Date(., format = "%Y-%m-%d") )) %>%
mutate(CA19.9=as.double(replace(dat0$CA19.9, dat0$CA19.9 == "<1.0", 1)))
summary(dat1)
-결측(대문자-"NA")으로-처리해보자." aria-hidden="true">문자열"."과 "na"이가 들어가있는 숫자형 변수 -> 결측(대문자 "NA")으로 처리해보자.
dat0$adc_decrease
dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.) %>%
mutate_at(vars(OP_date, Recur_date),
list(~as.Date(., format = "%Y-%m-%d") )) %>%
mutate(CA19.9=as.double(replace(dat0$CA19.9, dat0$CA19.9 == "<1.0", 1)),
adc_decrease=as.double(replace(
adc_decrease, adc_decrease=="na" | adc_decrease==".", NA)))
summary(dat1)
dat0$volume_change
dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.) %>%
mutate_at(vars(OP_date, Recur_date),
list(~as.Date(., format = "%Y-%m-%d") )) %>%
mutate(CA19.9=as.double(replace(dat0$CA19.9, dat0$CA19.9 == "<1.0", 1)),
adc_decrease=as.double(replace(
adc_decrease, adc_decrease=="na" | adc_decrease==".", NA)),
# 마지막엔 더블형변환해야하니 as.double부터
volume_change=as.double(replace(volume_change,
volume_change=="n/a", NA)))
범주형인데, 연속형으로 표기되어있는 -> mutate_at</h4>
- local_6m , sex?
</div>
</div>
</div>
dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.) %>%
mutate_at(vars(OP_date, Recur_date),
list(~as.Date(., format = "%Y-%m-%d") )) %>%
mutate(CA19.9=as.double(replace(dat0$CA19.9, dat0$CA19.9 == "<1.0", 1)),
adc_decrease=as.double(replace(
adc_decrease, adc_decrease=="na" | adc_decrease==".", NA)),
# 마지막엔 더블형변환해야하니 as.double부터
volume_change=as.double(replace(volume_change,
volume_change=="n/a", NA))) %>%
mutate_at(vars(sex, Recur, local_6m, TNM, CCI),
list(~as.factor(.)))
summary(dat1)
hist(dat1$CA19.9)
hist( log)
dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.) %>%
mutate_at(vars(OP_date, Recur_date),
list(~as.Date(., format = "%Y-%m-%d") )) %>%
mutate(CA19.9=as.double(replace(dat0$CA19.9, dat0$CA19.9 == "<1.0", 1)),
adc_decrease=as.double(replace(
adc_decrease, adc_decrease=="na" | adc_decrease==".", NA)),
volume_change=as.double(replace(volume_change,
volume_change=="n/a", NA)),
# 여기(mutate)다 추가
log.CA19.9 = log(CA19.9),
log.CEA=log(CEA)
# 위에다 추가
) %>%
mutate_at(vars(sex, Recur, local_6m, TNM, CCI),
list(~as.factor(.)))
dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.) %>%
mutate_at(vars(OP_date, Recur_date),
list(~as.Date(., format = "%Y-%m-%d") )) %>%
mutate(CA19.9=as.double(replace(dat0$CA19.9, dat0$CA19.9 == "<1.0", 1)),
adc_decrease=as.double(replace(
adc_decrease, adc_decrease=="na" | adc_decrease==".", NA)),
volume_change=as.double(replace(volume_change,
volume_change=="n/a", NA)),
log.CA19.9 = log(CA19.9),
log.CEA=log(CEA),
CA19.9.group=ifelse(CA19.9>37, 1, 0), # 칼럼 생성
) %>%
mutate_at(vars(sex, Recur, local_6m, TNM, CCI,
CA19.9 # factor화 추가
),
list(~as.factor(.)))
dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.) %>%
mutate_at(vars(OP_date, Recur_date),
list(~as.Date(., format = "%Y-%m-%d") )) %>%
mutate(CA19.9=as.double(replace(dat0$CA19.9, dat0$CA19.9 == "<1.0", 1)),
adc_decrease=as.double(replace(
adc_decrease, adc_decrease=="na" | adc_decrease==".", NA)),
volume_change=as.double(replace(volume_change,
volume_change=="n/a", NA)),
log.CA19.9 = log(CA19.9),
log.CEA=log(CEA),
CA19.9.group=ifelse(CA19.9>37, 1, 0),
TNM.b = mapvalues(TNM, from=c(1,2,3,4,5),
to=c("1or2", "1or2", "3", "4", "5")), # 카테고리 합치기
) %>%
mutate_at(vars(sex, Recur, local_6m, TNM, CCI,
CA19.9, TNM.b # 추가
),
list(~as.factor(.)))
dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.) %>%
mutate_at(vars(OP_date, Recur_date),
list(~as.Date(., format = "%Y-%m-%d") )) %>%
mutate(CA19.9=as.double(replace(dat0$CA19.9, dat0$CA19.9 == "<1.0", 1)),
adc_decrease=as.double(replace(
adc_decrease, adc_decrease=="na" | adc_decrease==".", NA)),
volume_change=as.double(replace(volume_change,
volume_change=="n/a", NA)),
log.CA19.9 = log(CA19.9),
log.CEA=log(CEA),
CA19.9.group=ifelse(CA19.9>37, 1, 0),
TNM.b = mapvalues(TNM, from=c(1,2,3,4,5),
to=c("1or2", "1or2", "3", "4", "5")),
CCI.b = mapvalues(CCI, from=c(0,1,2,3,4,6),
to=c("0or1", "0or1", "2", "3", ">=4", ">=4")),
) %>%
mutate_at(vars(sex, Recur, local_6m, TNM, CCI,
CA19.9, TNM.b # 추가
),
list(~as.factor(.)))
# as.factor가 아닌 factor로 어떤 순서대로 카테고리를 하고싶은지 인식시켜준다
# 아래에서 as.factor부분에서 빼준다.
library("tableone")
var1<-c("Recur", "RFS", "local_6m", "age", "sex", "CA19.9", "log.CA19.9", "CA19.9.group", "CEA", "log.CEA",
"TNM", "TNM.b", "CCI", "CCI.b", "adc_decrease", "adc_5_decrease", "volume_change",
"apt_1_mean", "apt_1_95p", "adc_1_mean", "adc_1_5p")
tobj<-CreateTableOne(vars = var1, data=dat1)
summary(tobj)
# 범주형 변수들의 요약통계량이 나온다. 갯수와 %
print(tobj)
print(tobj, showAllLevels = T)
# 하지만 치우친 [기울어진 분포]의 연속형변수는 [ median, IQR ]를 뽑아야한다.
print(tobj, showAllLevels = T, nonnormal = c("CA19.9", "CEA"))
?CreateTableOne
# createtableone에서 strata에 그룹을 나눈 칼럼명
options(width = 300)
tobj2<-CreateTableOne(vars = var1, data=dat1, strata="CA19.9.group")
print(tobj2)
</div>
같은작업들>
dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.) %>%
mutate_at(vars(OP_date, Recur_date),
list(~as.Date(., format = "%Y-%m-%d") )) %>%
mutate(CA19.9=as.double(replace(dat0$CA19.9, dat0$CA19.9 == "<1.0", 1)),
adc_decrease=as.double(replace(
adc_decrease, adc_decrease=="na" | adc_decrease==".", NA)),
# 마지막엔 더블형변환해야하니 as.double부터
volume_change=as.double(replace(volume_change,
volume_change=="n/a", NA))) %>%
mutate_at(vars(sex, Recur, local_6m, TNM, CCI),
list(~as.factor(.)))
summary(dat1)
hist(dat1$CA19.9)
hist( log)
dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.) %>%
mutate_at(vars(OP_date, Recur_date),
list(~as.Date(., format = "%Y-%m-%d") )) %>%
mutate(CA19.9=as.double(replace(dat0$CA19.9, dat0$CA19.9 == "<1.0", 1)),
adc_decrease=as.double(replace(
adc_decrease, adc_decrease=="na" | adc_decrease==".", NA)),
volume_change=as.double(replace(volume_change,
volume_change=="n/a", NA)),
# 여기(mutate)다 추가
log.CA19.9 = log(CA19.9),
log.CEA=log(CEA)
# 위에다 추가
) %>%
mutate_at(vars(sex, Recur, local_6m, TNM, CCI),
list(~as.factor(.)))
dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.) %>%
mutate_at(vars(OP_date, Recur_date),
list(~as.Date(., format = "%Y-%m-%d") )) %>%
mutate(CA19.9=as.double(replace(dat0$CA19.9, dat0$CA19.9 == "<1.0", 1)),
adc_decrease=as.double(replace(
adc_decrease, adc_decrease=="na" | adc_decrease==".", NA)),
volume_change=as.double(replace(volume_change,
volume_change=="n/a", NA)),
log.CA19.9 = log(CA19.9),
log.CEA=log(CEA),
CA19.9.group=ifelse(CA19.9>37, 1, 0), # 칼럼 생성
) %>%
mutate_at(vars(sex, Recur, local_6m, TNM, CCI,
CA19.9 # factor화 추가
),
list(~as.factor(.)))
dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.) %>%
mutate_at(vars(OP_date, Recur_date),
list(~as.Date(., format = "%Y-%m-%d") )) %>%
mutate(CA19.9=as.double(replace(dat0$CA19.9, dat0$CA19.9 == "<1.0", 1)),
adc_decrease=as.double(replace(
adc_decrease, adc_decrease=="na" | adc_decrease==".", NA)),
volume_change=as.double(replace(volume_change,
volume_change=="n/a", NA)),
log.CA19.9 = log(CA19.9),
log.CEA=log(CEA),
CA19.9.group=ifelse(CA19.9>37, 1, 0),
TNM.b = mapvalues(TNM, from=c(1,2,3,4,5),
to=c("1or2", "1or2", "3", "4", "5")), # 카테고리 합치기
) %>%
mutate_at(vars(sex, Recur, local_6m, TNM, CCI,
CA19.9, TNM.b # 추가
),
list(~as.factor(.)))
dat1 <- dat0 %>% rename(Recur=Recur..1..Recur..0..Censored.) %>%
mutate_at(vars(OP_date, Recur_date),
list(~as.Date(., format = "%Y-%m-%d") )) %>%
mutate(CA19.9=as.double(replace(dat0$CA19.9, dat0$CA19.9 == "<1.0", 1)),
adc_decrease=as.double(replace(
adc_decrease, adc_decrease=="na" | adc_decrease==".", NA)),
volume_change=as.double(replace(volume_change,
volume_change=="n/a", NA)),
log.CA19.9 = log(CA19.9),
log.CEA=log(CEA),
CA19.9.group=ifelse(CA19.9>37, 1, 0),
TNM.b = mapvalues(TNM, from=c(1,2,3,4,5),
to=c("1or2", "1or2", "3", "4", "5")),
CCI.b = mapvalues(CCI, from=c(0,1,2,3,4,6),
to=c("0or1", "0or1", "2", "3", ">=4", ">=4")),
) %>%
mutate_at(vars(sex, Recur, local_6m, TNM, CCI,
CA19.9, TNM.b # 추가
),
list(~as.factor(.)))
# as.factor가 아닌 factor로 어떤 순서대로 카테고리를 하고싶은지 인식시켜준다
# 아래에서 as.factor부분에서 빼준다.
library("tableone")
var1<-c("Recur", "RFS", "local_6m", "age", "sex", "CA19.9", "log.CA19.9", "CA19.9.group", "CEA", "log.CEA",
"TNM", "TNM.b", "CCI", "CCI.b", "adc_decrease", "adc_5_decrease", "volume_change",
"apt_1_mean", "apt_1_95p", "adc_1_mean", "adc_1_5p")
tobj<-CreateTableOne(vars = var1, data=dat1)
summary(tobj)
# 범주형 변수들의 요약통계량이 나온다. 갯수와 %
print(tobj)
print(tobj, showAllLevels = T)
# 하지만 치우친 [기울어진 분포]의 연속형변수는 [ median, IQR ]를 뽑아야한다.
print(tobj, showAllLevels = T, nonnormal = c("CA19.9", "CEA"))
?CreateTableOne
# createtableone에서 strata에 그룹을 나눈 칼럼명
options(width = 300)
tobj2<-CreateTableOne(vars = var1, data=dat1, strata="CA19.9.group")
print(tobj2)